Direkt-siRNA™
Enabling technology for systematic knock-down of gene expression and for automated, high throughput gene function analysis in mammalian cells.
Among the 30,000-40,000 human genes discovered as a result of the recently completed human genome project, the function of at least half of them remains unknown. Deciphering the functions of these genes becomes the next frontier for biomedical research and drug discovery efforts. The most straightforward way of probing the function of a gene is to somehow get rid of the gene (or the gene product) of interest and to observe what happens without this gene (or its product). Many technologies have been used to delete genes (gene knock out), to suppress gene expression (e.g. antisense and ribozymes) or to inhibit the activity of gene products (e.g. dominant negative mutants). Except for targeted gene knock-out by homologous recombinantion, which is costly and slow, these approaches are useful to a very limited degree. The recently discovered small interfering RNAs (siRNA) technology, however, is revolutionizing the systematic analysis of gene functions with its promise of suppressing the expression of any gene in vertebrate cells.
RNA interference (RNAi) refers to the phenomenon of double-stranded RNA (dsRNA)-mediated, post-transcription mRNA degradation. It’s is triggered by dsRNA and mediated by small interference RNA duplexes (siRNA) of 21-23 nucleotides. Longer dsRNAs (500-1000bp) worked very well in many evolutionarily diverse organisms including plants, insects, fungi and metazoans. However, longer dsRNAs do not work in mammals because they provokes a strong cytotoxic response, possibly due to the non-specific effects on gene expression that result from activation of the interferon (IFN)-related pathways by long (>30bp) dsRNA.
Dlbashir et. Al, (2001) first demonstrated that synthesized short dsRNAs of ~21 bp can efficiently mediate sequence-specific mRNA degradation and suppress gene expression in cultured mammalian cells without triggering non-specific mRNA degradation as do longer dsRNAs. This report fueled a surge in the interest in using siRNAs in mammalian cells for gene function analysis and as antiviral reagents and resulted in a flood of papers describing the use of chemically synthesized dsRNAs to suppress various gene expressions in many common cell lines. Because chemically synthesized siRNAs are expersive, several groups have devised strategies to synthesize short RNAs using in vitro transcription methods directly (Donze and Picard, 2002) or in combination with RNAse III hydrolysis (Yang et al., 2002). A little later, nine papers appeared in three months and demonstrated that siRNAs expressed in mammalian cells from plasmid templates can silence gene expression as effectively as do synthetic siRNAs. In mammalian cells, RNA polymerase III is responsible for the synthesis of many small heterogeneous nuclear RNAs involved in RNA splicing and other processes. Not surprisingly, all these reports used pol III promoters, such as H1 RNA and U6 snRNA promoter, to drive the synthesis of siRNAs or self-complimentary hairpins.
To overcome the limitations of using chemically synthesized double strand siRNAs, Biomyx developed a series of proprietary DNA oligo-based means of silencing gene expressions in mammalian cells. pH1-siRNA and pU6-siRNA (siRNA Vectors) are mammalian expression vectors developed at Biomyx. A chosen sequence (from the targeted mRNA) cloned easily into the vectors as a inverted repeat under the promoter will be expressed in mammalian cells as a hairpin structure and processed in vivo to produce siRNA, which initiate the RNA interference effects. They offer researchers the choice of either one of the two most widely used promoters for expressing siRNA: the human H1 RNA promoter or the mouse U6 snRNA promoter. They are small in size and user-friendly. Both vectors have a G418 resistant gene expression cassette enabling the selection of stable cell lines expressing the desired siRNA. As compared with the use of synthetic or in vitro transcribed siRNAs, these DNA oligo-based approaches are cost efficient, more consistent and can be integrated into the genome to study long term effects of siRNAs.
The making of these vectors, however, is time consuming and hence limited our ability in exploring the full power of RNAi. It is generally believed that only one out of 4 siRNA sequences will be very effective in knocking down. To find an effective siRNA sequence for a given gene, many expression vectors need to be constructed, not mentioning the systematic knock-down of many genes simultaneusly. To meet the need of high throughput, systematic gene function studies, Biomyx developed the proprietary Direkt siRNATM technology that allows the generation, delivery and expression of many siRNAs at minimal cost. The technology will enable researchers to scan mRNA or untranslated sequences for effective siRNA sequences, or to study the effects of siRNAs for many genes in a matter of days, which is impractical with synthetic dsRNA or expression vector based methods.
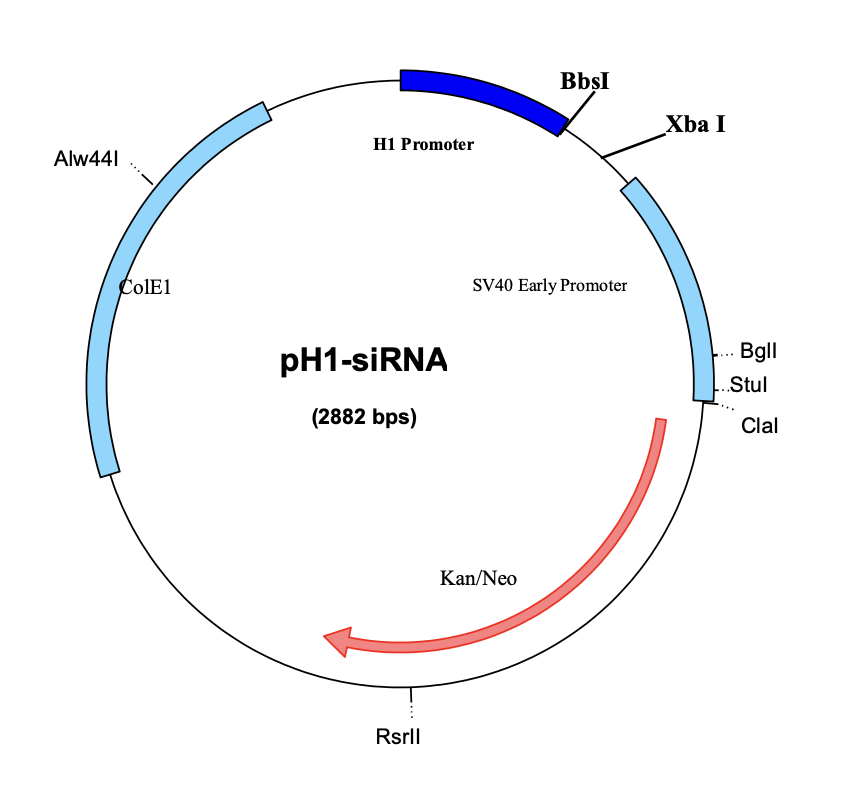
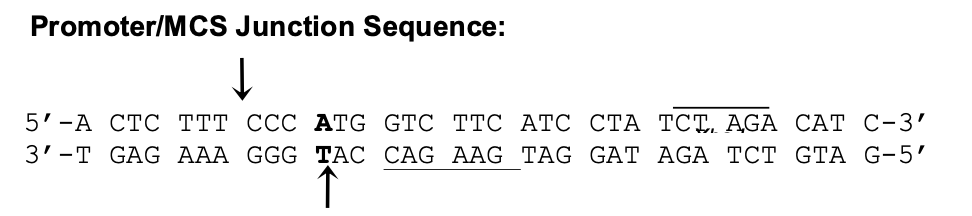
Circular Map of pU6-siRNA:
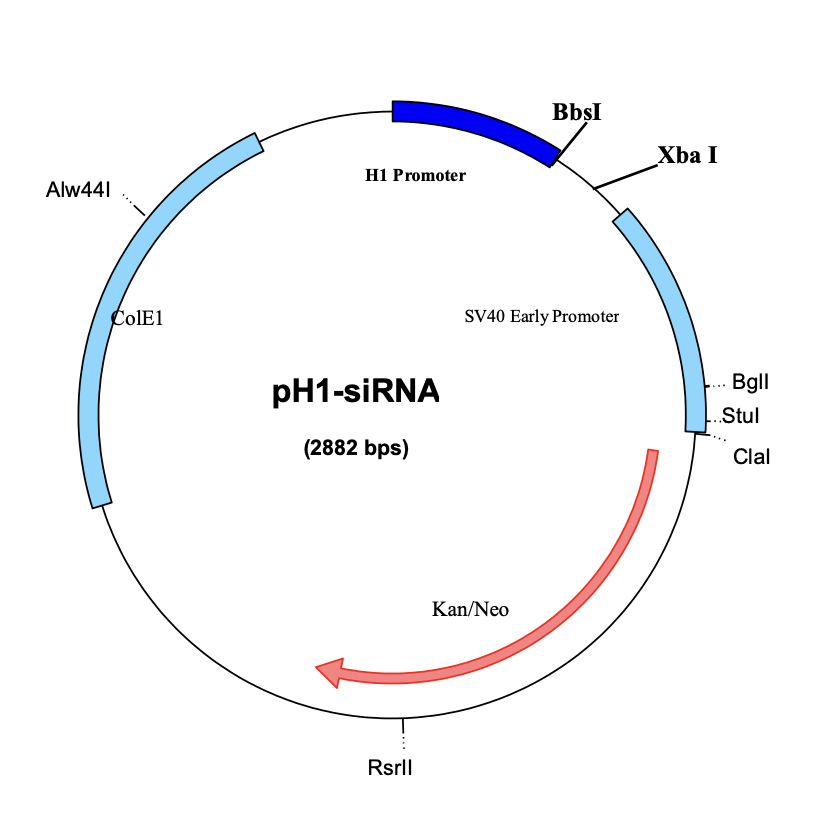
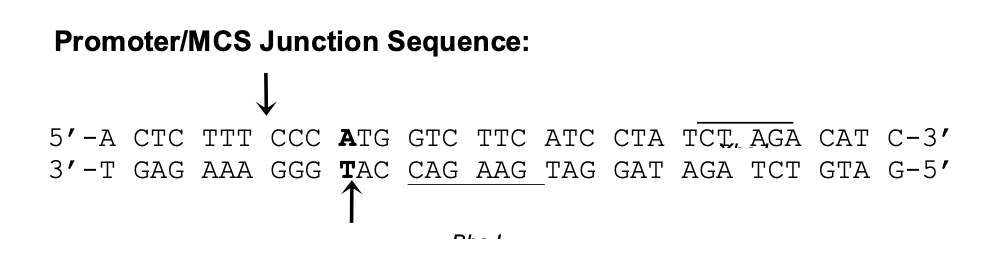
Circular Map of pH1-siRNA: